Molybdenum »
PDB 1aa6-1n61 »
1eoi »
Molybdenum in PDB 1eoi: Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate
Enzymatic activity of Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate
All present enzymatic activity of Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate:
3.1.3.2;
3.1.3.2;
Protein crystallography data
The structure of Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate, PDB code: 1eoi
was solved by
K.Ishikawa,
Y.Mihara,
K.Gondoh,
E.Suzuki,
Y.Asano,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 10.00 / 2.40 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.600, 86.600, 205.300, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 21.2 / 28 |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate
(pdb code 1eoi). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total 3 binding sites of Molybdenum where determined in the Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate, PDB code: 1eoi:
Jump to Molybdenum binding site number: 1; 2; 3;
In total 3 binding sites of Molybdenum where determined in the Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate, PDB code: 1eoi:
Jump to Molybdenum binding site number: 1; 2; 3;
Molybdenum binding site 1 out of 3 in 1eoi
Go back to
Molybdenum binding site 1 out
of 3 in the Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate
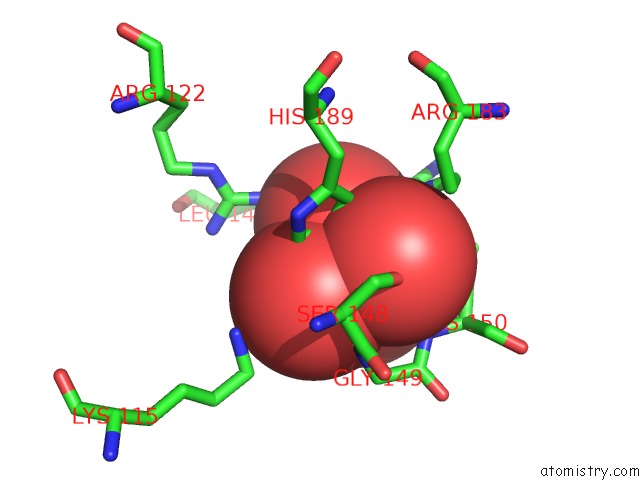
Mono view
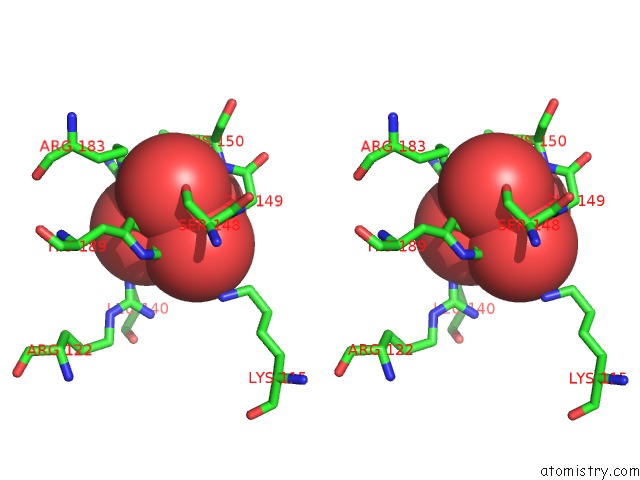
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate within 5.0Å range:
|
Molybdenum binding site 2 out of 3 in 1eoi
Go back to
Molybdenum binding site 2 out
of 3 in the Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate
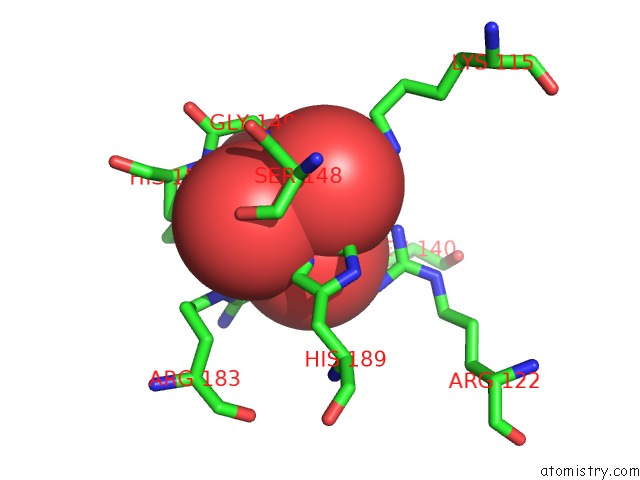
Mono view
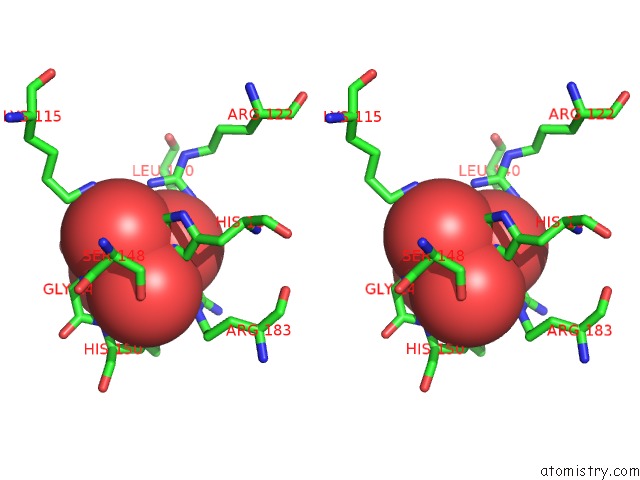
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 2 of Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate within 5.0Å range:
|
Molybdenum binding site 3 out of 3 in 1eoi
Go back to
Molybdenum binding site 3 out
of 3 in the Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate
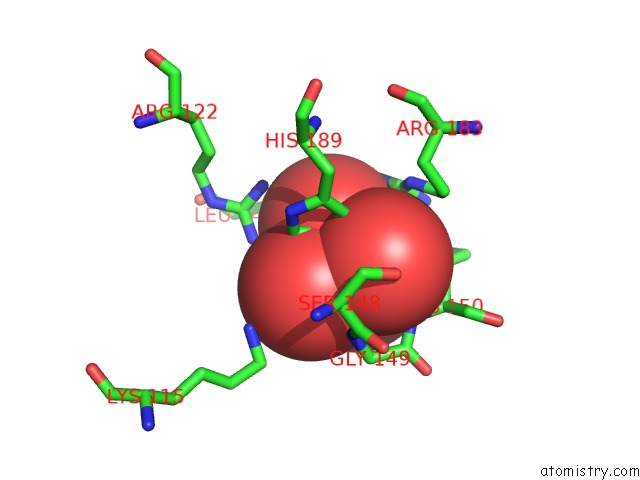
Mono view
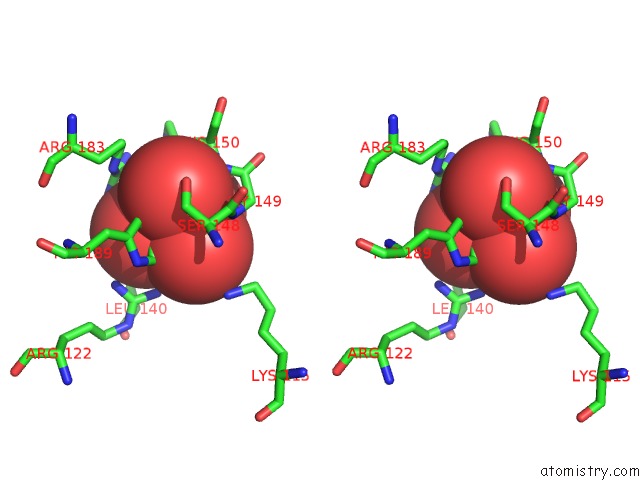
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 3 of Crystal Structure of Acid Phosphatase From Escherichia Blattae Complexed with the Transition State Analog Molybdate within 5.0Å range:
|
Reference:
K.Ishikawa,
Y.Mihara,
K.Gondoh,
E.Suzuki,
Y.Asano.
X-Ray Structures of A Novel Acid Phosphatase From Escherichia Blattae and Its Complex with the Transition-State Analog Molybdate. Embo J. V. 19 2412 2000.
ISSN: ISSN 0261-4189
PubMed: 10835340
DOI: 10.1093/EMBOJ/19.11.2412
Page generated: Sun Aug 17 02:51:27 2025
ISSN: ISSN 0261-4189
PubMed: 10835340
DOI: 10.1093/EMBOJ/19.11.2412
Last articles
Na in 2WHJNa in 2WGD
Na in 2WGE
Na in 2WG4
Na in 2WG9
Na in 2WFT
Na in 2WFX
Na in 2WG8
Na in 2WG7
Na in 2WDV