Molybdenum »
PDB 1aa6-1n61 »
1kqf »
Molybdenum in PDB 1kqf: Formate Dehydrogenase N From E. Coli
Enzymatic activity of Formate Dehydrogenase N From E. Coli
All present enzymatic activity of Formate Dehydrogenase N From E. Coli:
1.2.1.2;
1.2.1.2;
Protein crystallography data
The structure of Formate Dehydrogenase N From E. Coli, PDB code: 1kqf
was solved by
M.Jormakka,
S.Tornroth,
B.Byrne,
S.Iwata,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.00 / 1.60 |
| Space group | P 21 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 203.000, 203.000, 203.000, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.7 / 19.5 |
Other elements in 1kqf:
The structure of Formate Dehydrogenase N From E. Coli also contains other interesting chemical elements:
| Iron | (Fe) | 22 atoms |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Formate Dehydrogenase N From E. Coli
(pdb code 1kqf). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total only one binding site of Molybdenum was determined in the Formate Dehydrogenase N From E. Coli, PDB code: 1kqf:
In total only one binding site of Molybdenum was determined in the Formate Dehydrogenase N From E. Coli, PDB code: 1kqf:
Molybdenum binding site 1 out of 1 in 1kqf
Go back to
Molybdenum binding site 1 out
of 1 in the Formate Dehydrogenase N From E. Coli
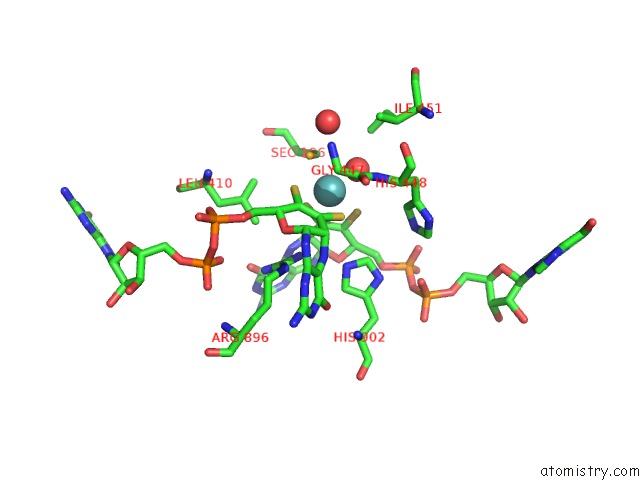
Mono view
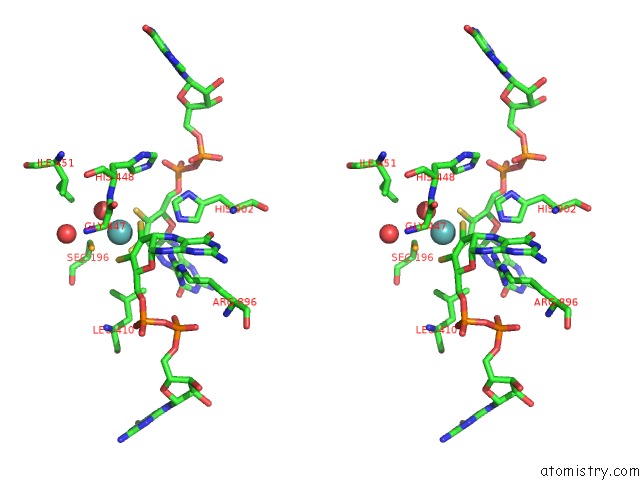
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Formate Dehydrogenase N From E. Coli within 5.0Å range:
|
Reference:
M.Jormakka,
S.Tornroth,
B.Byrne,
S.Iwata.
Molecular Basis of Proton Motive Force Generation: Structure of Formate Dehydrogenase-N. Science V. 295 1863 2002.
ISSN: ISSN 0036-8075
PubMed: 11884747
DOI: 10.1126/SCIENCE.1068186
Page generated: Sun Aug 17 02:55:35 2025
ISSN: ISSN 0036-8075
PubMed: 11884747
DOI: 10.1126/SCIENCE.1068186
Last articles
Na in 1VI6Na in 1VKG
Na in 1VMJ
Na in 1VMH
Na in 1VMF
Na in 1VLM
Na in 1VK1
Na in 1VIZ
Na in 1VEL
Na in 1VE8