Molybdenum »
PDB 3fc4-4c7z »
3r18 »
Molybdenum in PDB 3r18: Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity
Enzymatic activity of Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity
All present enzymatic activity of Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity:
1.8.3.1;
1.8.3.1;
Protein crystallography data
The structure of Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity, PDB code: 3r18
was solved by
J.A.Qiu,
H.L.Wilson,
K.V.Rajagopalan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.51 / 2.40 |
| Space group | I 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 85.623, 85.623, 153.371, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.5 / 20.7 |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity
(pdb code 3r18). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total only one binding site of Molybdenum was determined in the Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity, PDB code: 3r18:
In total only one binding site of Molybdenum was determined in the Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity, PDB code: 3r18:
Molybdenum binding site 1 out of 1 in 3r18
Go back to
Molybdenum binding site 1 out
of 1 in the Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity
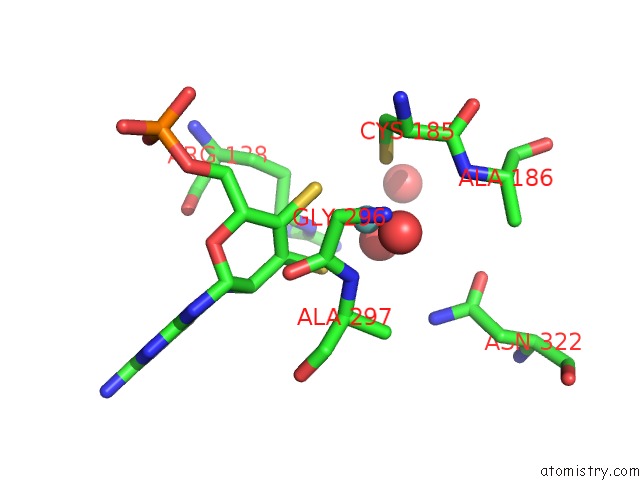
Mono view
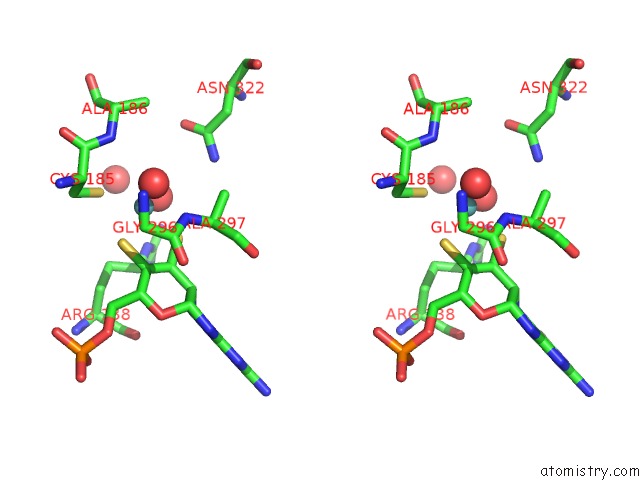
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Chicken Sulfite Oxidase Double Mutant with Altered Activity and Substrate Affinity within 5.0Å range:
|
Reference:
J.A.Qiu,
H.L.Wilson,
K.V.Rajagopalan.
Structure-Based Alteration of Substrate Specificity and Catalytic Activity of Sulfite Oxidase From Sulfite Oxidation to Nitrate Reduction. Biochemistry V. 51 1134 2012.
ISSN: ISSN 0006-2960
PubMed: 22263579
DOI: 10.1021/BI201206V
Page generated: Sun Oct 6 15:56:53 2024
ISSN: ISSN 0006-2960
PubMed: 22263579
DOI: 10.1021/BI201206V
Last articles
Cl in 7SUJCl in 7SUT
Cl in 7SUI
Cl in 7SUH
Cl in 7SQE
Cl in 7STV
Cl in 7STT
Cl in 7STQ
Cl in 7SSF
Cl in 7SSB