Molybdenum »
PDB 4c80-5koj »
5chc »
Molybdenum in PDB 5chc: Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps
Protein crystallography data
The structure of Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps, PDB code: 5chc
was solved by
C.-L.Tsai,
J.A.Tainer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.52 / 2.38 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 133.926, 176.022, 193.691, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 22.7 |
Other elements in 5chc:
The structure of Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
| Iron | (Fe) | 57 atoms |
| Sodium | (Na) | 8 atoms |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps
(pdb code 5chc). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total 3 binding sites of Molybdenum where determined in the Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps, PDB code: 5chc:
Jump to Molybdenum binding site number: 1; 2; 3;
In total 3 binding sites of Molybdenum where determined in the Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps, PDB code: 5chc:
Jump to Molybdenum binding site number: 1; 2; 3;
Molybdenum binding site 1 out of 3 in 5chc
Go back to
Molybdenum binding site 1 out
of 3 in the Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps
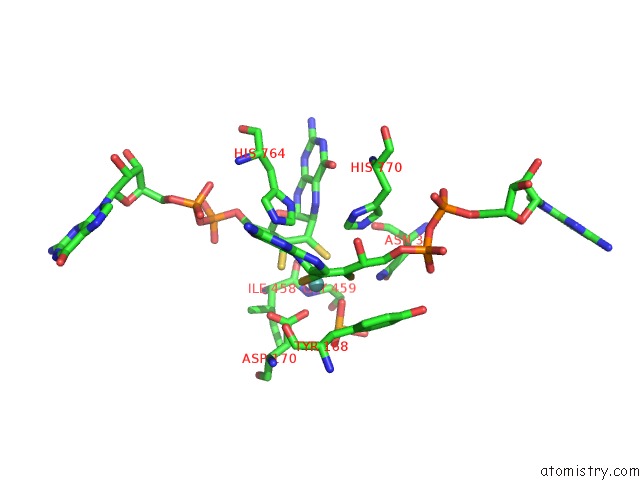
Mono view
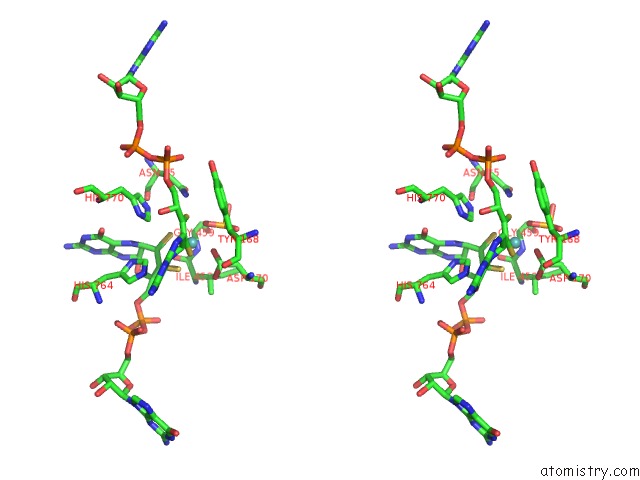
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps within 5.0Å range:
|
Molybdenum binding site 2 out of 3 in 5chc
Go back to
Molybdenum binding site 2 out
of 3 in the Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps
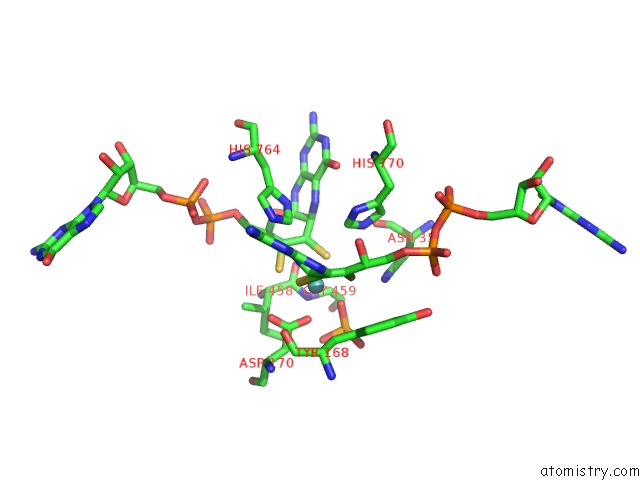
Mono view
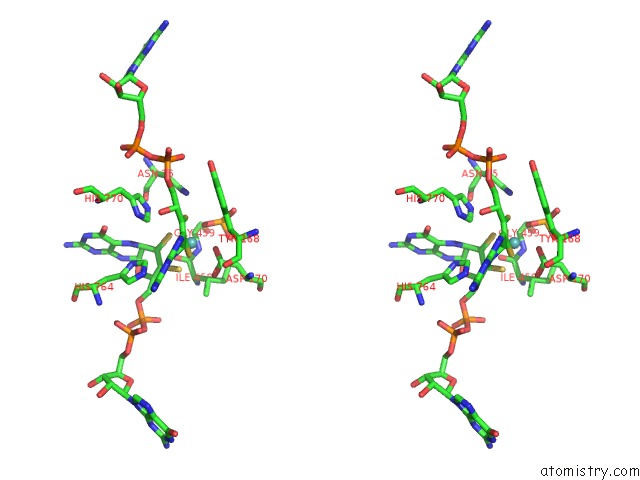
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 2 of Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps within 5.0Å range:
|
Molybdenum binding site 3 out of 3 in 5chc
Go back to
Molybdenum binding site 3 out
of 3 in the Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 3 of Crystal Structure of the Perchlorate Reductase Pcrab - Substrate Analog SEO3 Bound - From Azospira Suillum Ps within 5.0Å range:
|
Reference:
M.D.Youngblut,
C.L.Tsai,
I.C.Clark,
H.K.Carlson,
A.P.Maglaqui,
P.S.Gau-Pan,
S.A.Redford,
A.Wong,
J.A.Tainer,
J.D.Coates.
Perchlorate Reductase Is Distinguished By Active Site Aromatic Gate Residues. J.Biol.Chem. V. 291 9190 2016.
ISSN: ESSN 1083-351X
PubMed: 26940877
DOI: 10.1074/JBC.M116.714618
Page generated: Sun Oct 6 16:20:13 2024
ISSN: ESSN 1083-351X
PubMed: 26940877
DOI: 10.1074/JBC.M116.714618
Last articles
I in 4LDVI in 4LBR
I in 4LB3
I in 4LAZ
I in 4KSZ
I in 4L6C
I in 4K1C
I in 4KQV
I in 4KRW
I in 4KRL