Molybdenum »
PDB 4c80-5koj »
5k3x »
Molybdenum in PDB 5k3x: Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
Enzymatic activity of Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
All present enzymatic activity of Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti:
1.8.3.1;
1.8.3.1;
Protein crystallography data
The structure of Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti, PDB code: 5k3x
was solved by
M.Lee,
A.Mcgrath,
M.Maher,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 72.10 / 1.60 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.470, 95.720, 109.620, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.5 / 18 |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
(pdb code 5k3x). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total 2 binding sites of Molybdenum where determined in the Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti, PDB code: 5k3x:
Jump to Molybdenum binding site number: 1; 2;
In total 2 binding sites of Molybdenum where determined in the Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti, PDB code: 5k3x:
Jump to Molybdenum binding site number: 1; 2;
Molybdenum binding site 1 out of 2 in 5k3x
Go back to
Molybdenum binding site 1 out
of 2 in the Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
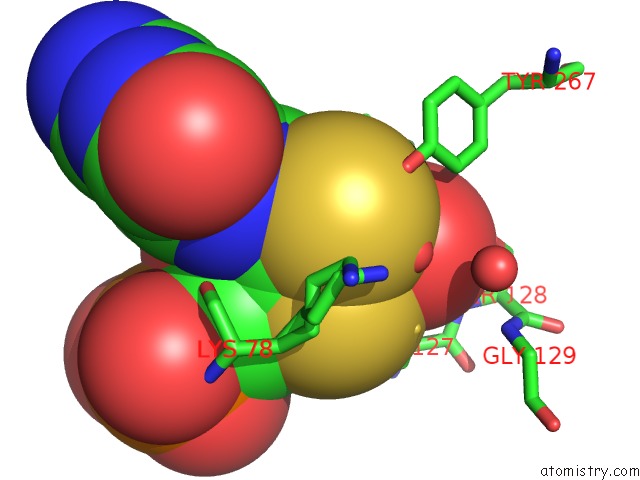
Mono view
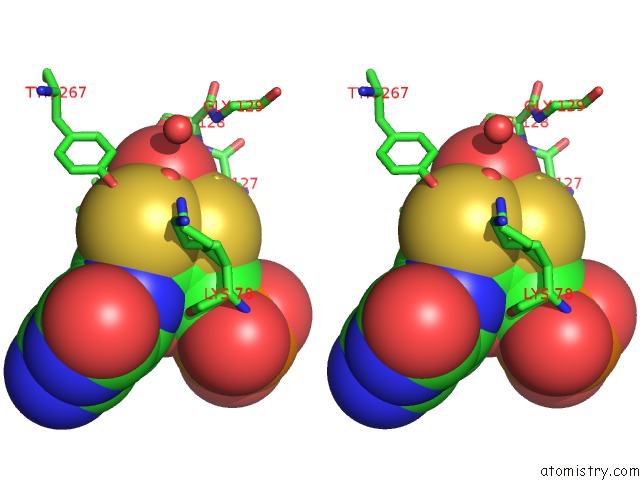
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti within 5.0Å range:
|
Molybdenum binding site 2 out of 2 in 5k3x
Go back to
Molybdenum binding site 2 out
of 2 in the Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti
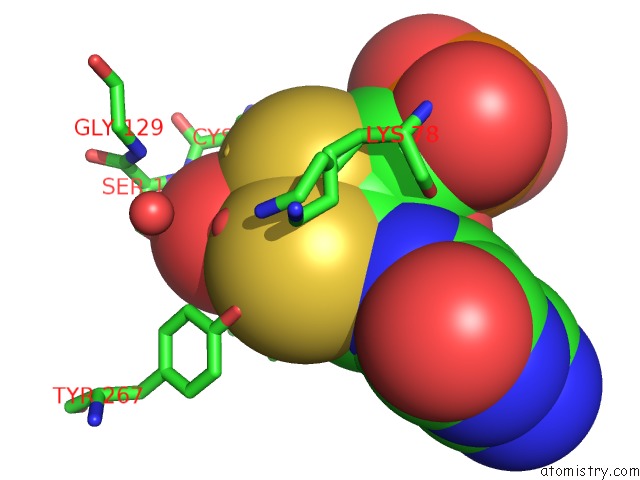
Mono view
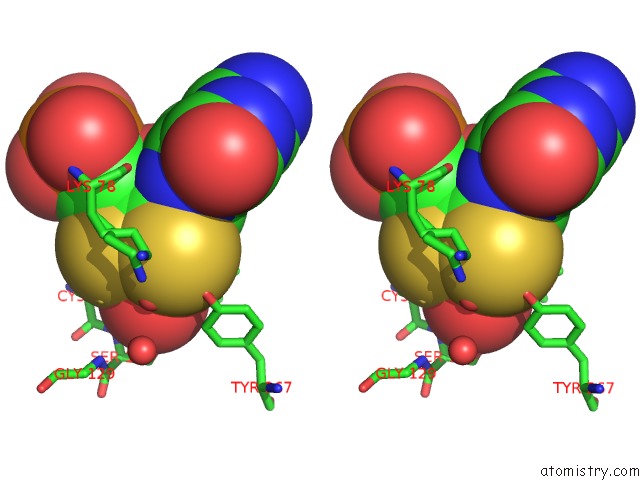
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 2 of Crystal Structure of the Sulfite Dehydrogenase, Sort R78K Mutant From Sinorhizobium Meliloti within 5.0Å range:
|
Reference:
J.C.Hsiao,
A.P.Mcgrath,
L.Kielmann,
P.Kalimuthu,
F.Darain,
P.V.Bernhardt,
J.Harmer,
M.Lee,
K.Meyers,
M.J.Maher,
U.Kappler.
The Central Active Site Arginine in Sulfite Oxidizing Enzymes Alters Kinetic Properties By Controlling Electron Transfer and Redox Interactions. Biochim. Biophys. Acta V.1859 19 2017.
ISSN: ISSN 0006-3002
PubMed: 28986298
DOI: 10.1016/J.BBABIO.2017.10.001
Page generated: Sun Aug 17 03:40:02 2025
ISSN: ISSN 0006-3002
PubMed: 28986298
DOI: 10.1016/J.BBABIO.2017.10.001
Last articles
Na in 1J9MNa in 1IYN
Na in 1IUE
Na in 1IP7
Na in 1IX0
Na in 1IP6
Na in 1IP3
Na in 1IOC
Na in 1IP5
Na in 1IP4