Molybdenum »
PDB 8cff-9d2c »
8e3u »
Molybdenum in PDB 8e3u: Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation
Enzymatic activity of Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation
All present enzymatic activity of Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation:
1.18.6.1;
1.18.6.1;
Protein crystallography data
The structure of Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation, PDB code: 8e3u
was solved by
H.L.Rutledge,
F.A.Tezcan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 79.89 / 1.99 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.259, 129.009, 107.76, 90, 109.22, 90 |
| R / Rfree (%) | 20.2 / 25.3 |
Other elements in 8e3u:
The structure of Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
| Iron | (Fe) | 30 atoms |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation
(pdb code 8e3u). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total 2 binding sites of Molybdenum where determined in the Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation, PDB code: 8e3u:
Jump to Molybdenum binding site number: 1; 2;
In total 2 binding sites of Molybdenum where determined in the Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation, PDB code: 8e3u:
Jump to Molybdenum binding site number: 1; 2;
Molybdenum binding site 1 out of 2 in 8e3u
Go back to
Molybdenum binding site 1 out
of 2 in the Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation
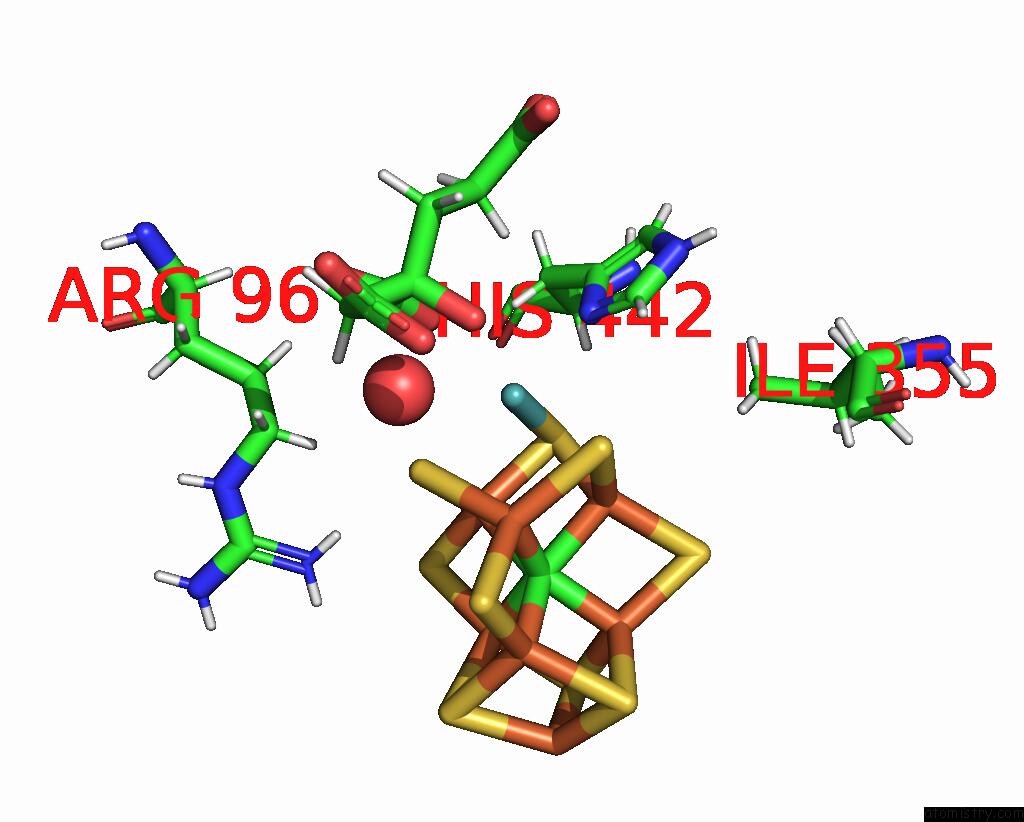
Mono view
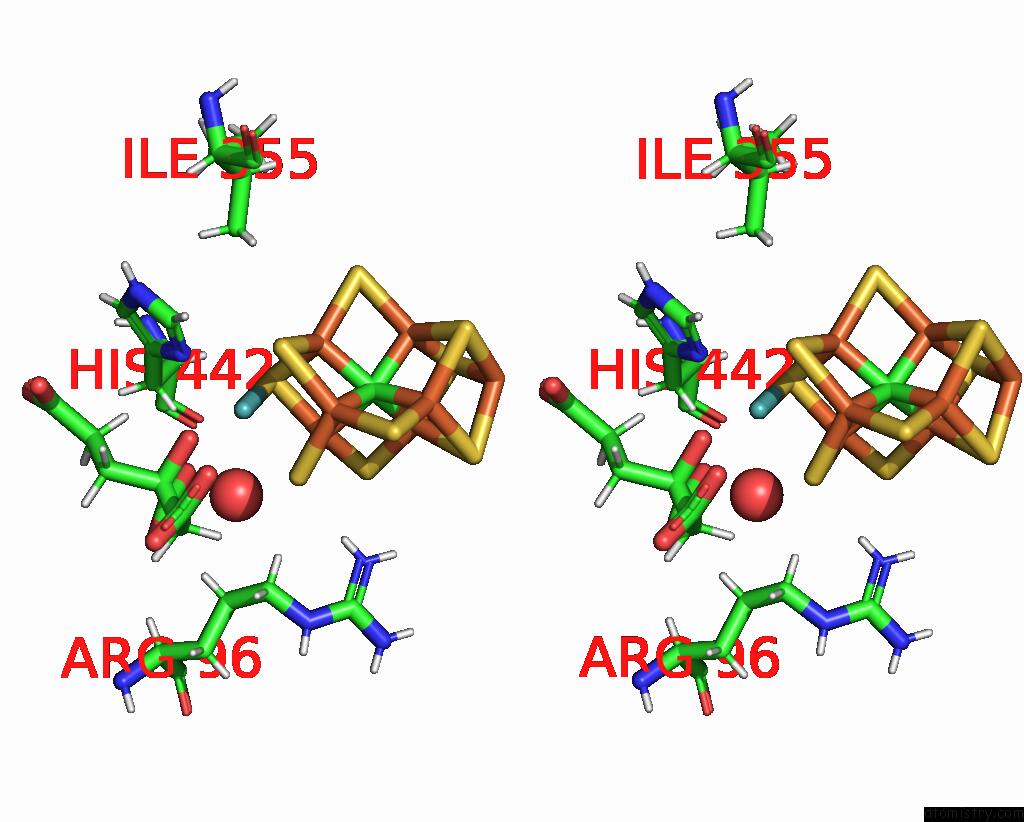
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation within 5.0Å range:
|
Molybdenum binding site 2 out of 2 in 8e3u
Go back to
Molybdenum binding site 2 out
of 2 in the Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation
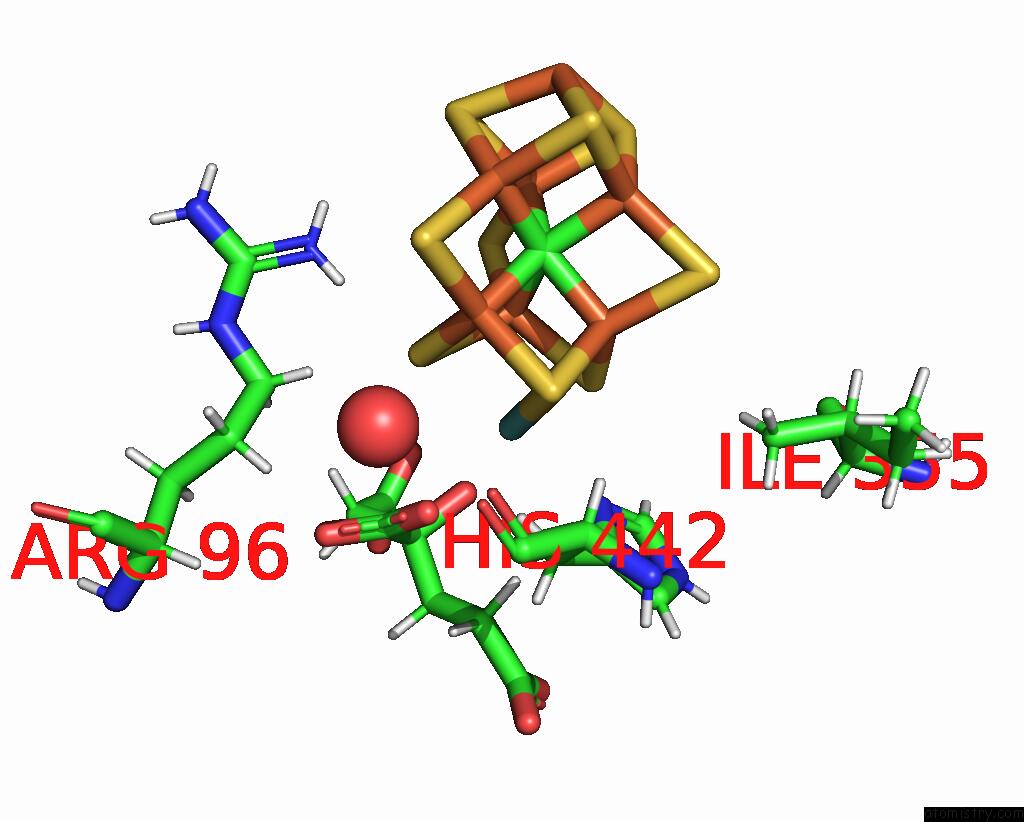
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 2 of Nickel-Reconstituted Nitrogenase Mofep Mutant S188A From Azotobacter Vinelandii After Ids Oxidation within 5.0Å range:
|
Reference:
H.L.Rutledge,
M.J.Field,
J.Rittle,
M.T.Green,
F.A.Tezcan.
Role of Serine Coordination in the Structural and Functional Protection of the Nitrogenase P-Cluster. J.Am.Chem.Soc. V. 144 22101 2022.
ISSN: ESSN 1520-5126
PubMed: 36445204
DOI: 10.1021/JACS.2C09480
Page generated: Sun Oct 6 17:41:55 2024
ISSN: ESSN 1520-5126
PubMed: 36445204
DOI: 10.1021/JACS.2C09480
Last articles
F in 4IQVF in 4IQW
F in 4IQT
F in 4IQU
F in 4INB
F in 4IKT
F in 4IJU
F in 4IKL
F in 4IJV
F in 4IKK