Molybdenum »
PDB 8cff-9cqy »
8eno »
Molybdenum in PDB 8eno: Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft
Enzymatic activity of Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft
All present enzymatic activity of Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft:
1.18.6.1;
1.18.6.1;
Other elements in 8eno:
The structure of Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft also contains other interesting chemical elements:
| Iron | (Fe) | 32 atoms |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft
(pdb code 8eno). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total 2 binding sites of Molybdenum where determined in the Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft, PDB code: 8eno:
Jump to Molybdenum binding site number: 1; 2;
In total 2 binding sites of Molybdenum where determined in the Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft, PDB code: 8eno:
Jump to Molybdenum binding site number: 1; 2;
Molybdenum binding site 1 out of 2 in 8eno
Go back to
Molybdenum binding site 1 out
of 2 in the Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft within 5.0Å range:
|
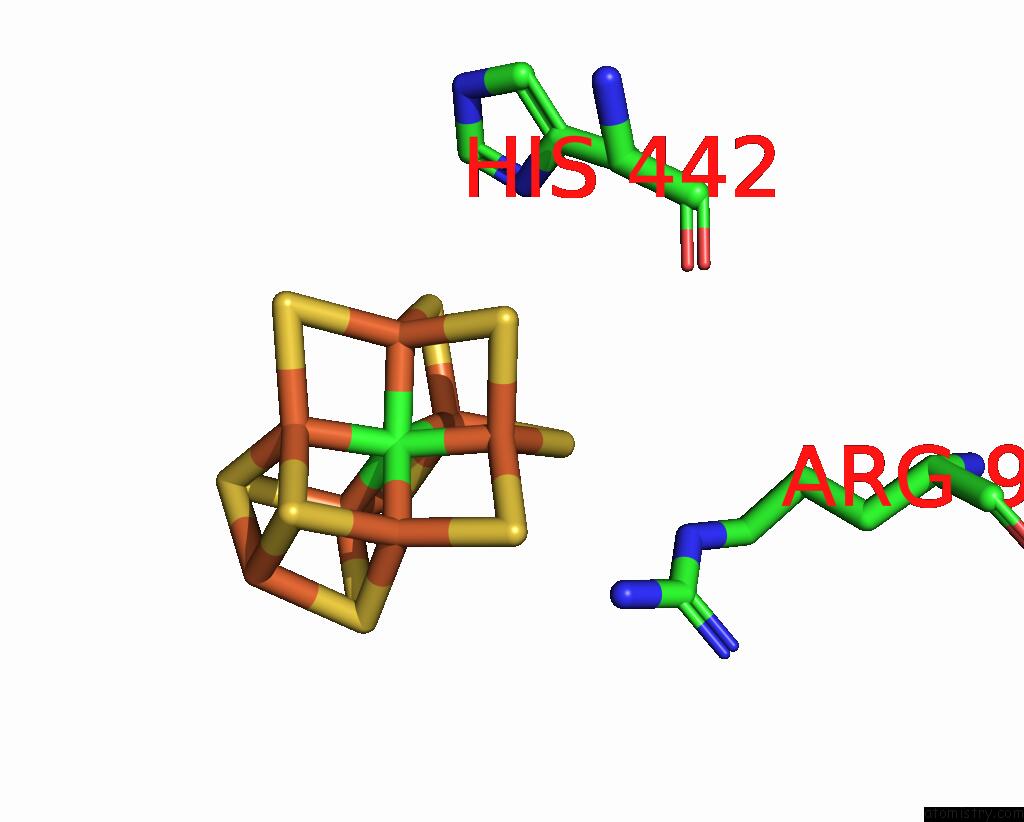
Molybdenum binding site 2 out of 2 in 8eno
Go back to
Molybdenum binding site 2 out
of 2 in the Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft
Mono view
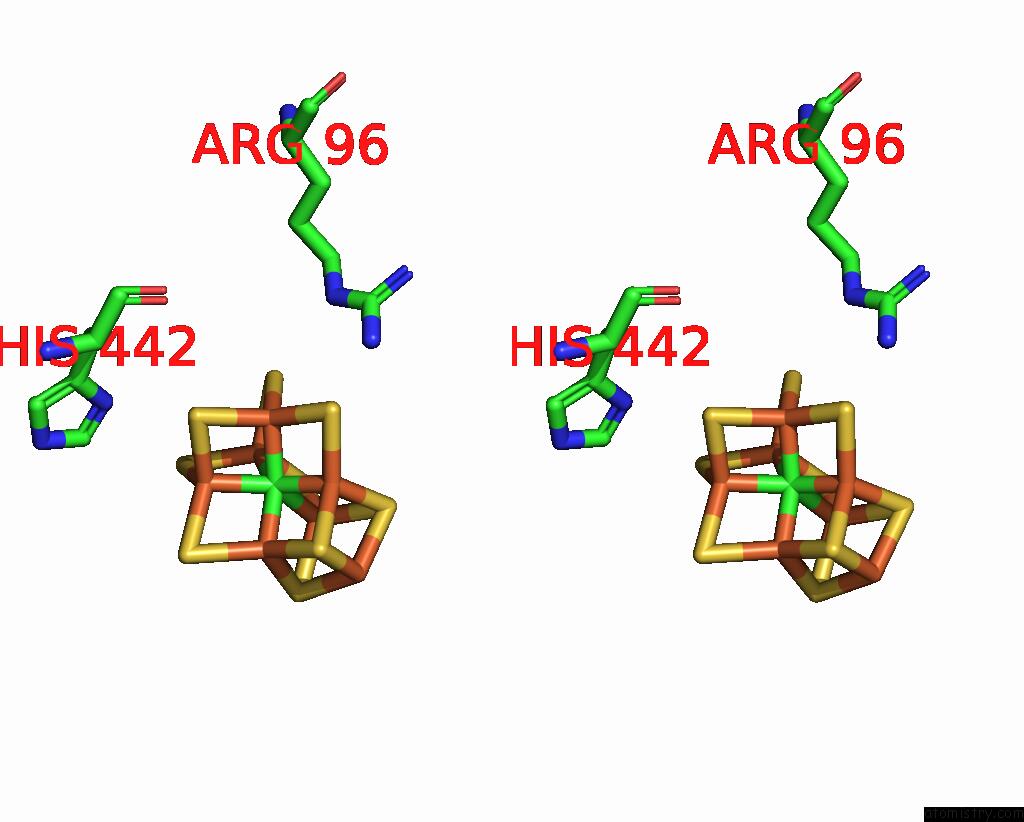
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 2 of Homocitrate-Deficient Nitrogenase Mofe-Protein From A. Vinelandii Nifv Knockout in Complex with Naft within 5.0Å range:
|
Reference:
R.A.Warmack,
A.O.Maggiolo,
A.Orta,
B.B.Wenke,
J.B.Howard,
D.C.Rees.
Structural Consequences of Turnover-Induced Homocitrate Loss in Nitrogenase. Nat Commun V. 14 1091 2023.
ISSN: ESSN 2041-1723
PubMed: 36841829
DOI: 10.1038/S41467-023-36636-4
Page generated: Sun Aug 17 04:33:39 2025
ISSN: ESSN 2041-1723
PubMed: 36841829
DOI: 10.1038/S41467-023-36636-4
Last articles
Na in 4ZM0Na in 4ZLA
Na in 4ZK6
Na in 4ZI6
Na in 4ZLS
Na in 4ZKE
Na in 4ZKY
Na in 4ZIP
Na in 4ZIX
Na in 4ZJU