Molybdenum »
PDB 8cff-9cqy »
8gy3 »
Molybdenum in PDB 8gy3: Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans
Enzymatic activity of Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans
All present enzymatic activity of Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans:
1.2.99.7;
1.2.99.7;
Other elements in 8gy3:
The structure of Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans also contains other interesting chemical elements:
| Iron | (Fe) | 7 atoms |
Molybdenum Binding Sites:
The binding sites of Molybdenum atom in the Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans
(pdb code 8gy3). This binding sites where shown within
5.0 Angstroms radius around Molybdenum atom.
In total only one binding site of Molybdenum was determined in the Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans, PDB code: 8gy3:
In total only one binding site of Molybdenum was determined in the Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans, PDB code: 8gy3:
Molybdenum binding site 1 out of 1 in 8gy3
Go back to
Molybdenum binding site 1 out
of 1 in the Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans

Mono view
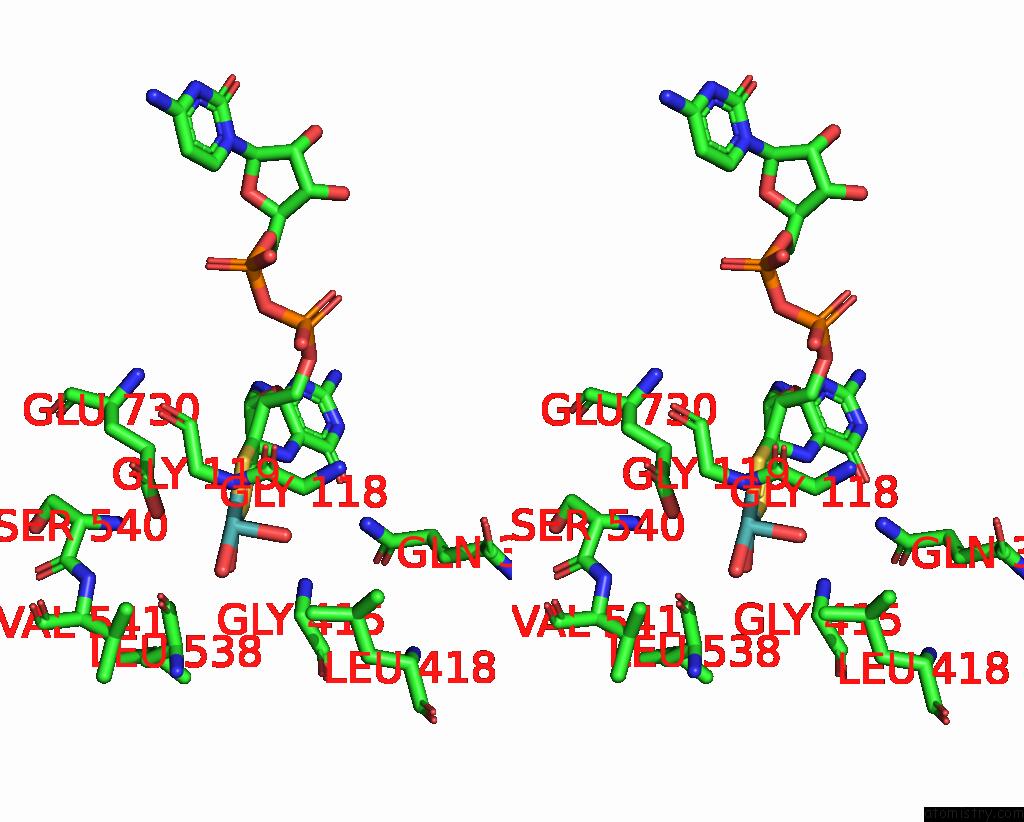
Stereo pair view

Mono view

Stereo pair view
A full contact list of Molybdenum with other atoms in the Mo binding
site number 1 of Cryo-Em Structure of Membrane-Bound Aldehyde Dehydrogenase From Gluconobacter Oxydans within 5.0Å range:
|
Reference:
T.Adachi,
T.Miyata,
F.Makino,
H.Tanaka,
K.Namba,
K.Kano,
K.Sowa,
Y.Kitazumi,
O.Shirai.
Experimental and Theoretical Insights Into Bienzymatic Cascade For Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases Acs Catalysis V. 13 7955 2023.
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.3C01962
Page generated: Sun Aug 17 04:33:42 2025
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.3C01962
Last articles
Na in 7DJLNa in 7DJC
Na in 7DJJ
Na in 7DJ2
Na in 7DJ1
Na in 7DII
Na in 7DIX
Na in 7DGC
Na in 7DER
Na in 7DFN